Chlamydial Infection: Chlamydia trachomatis
 Chlamydia trachomatis is a potent sexually transmitted disease, that can infect both Homo sapiens and amoeba (Oriel and Hayward, 1974). C. trachomatis is a coccoid shaped bacterium (figure 1) with gram-negative properties, as an obligate intracellular bacteria C. trachomatis acts as an extremely successful pathogen (Oriel and Hayward, 1974). C. trachomatis was initially thought to be a virus, because C. trachomatis is unable to carry out metabolic processes and has a deficiency in many biosynthetic pathways. This means, much like viruses they are completely dependent on the host supplying them with ATP, nutrients, amino acids and nucleoside triphosphates (Malhotra et al., 2013). However, unlike viruses, C. trachomatis possesses both DNA and RNA, possess their own ribosomes and replicate through binary fission, therefore they are known to be a bacterium (Becker et al., 1996). C. trachomatis is a powerful immunogen; meaning it is bound to components of the immune system, this stimulates both humeral and cell immune responses. The outcome associated with C. trachomatis really depends on the volume and interaction of cytokines secreted by lymphocytes (Mascellino, Boccia and Oliva, 2011).
Chlamydia trachomatis is a potent sexually transmitted disease, that can infect both Homo sapiens and amoeba (Oriel and Hayward, 1974). C. trachomatis is a coccoid shaped bacterium (figure 1) with gram-negative properties, as an obligate intracellular bacteria C. trachomatis acts as an extremely successful pathogen (Oriel and Hayward, 1974). C. trachomatis was initially thought to be a virus, because C. trachomatis is unable to carry out metabolic processes and has a deficiency in many biosynthetic pathways. This means, much like viruses they are completely dependent on the host supplying them with ATP, nutrients, amino acids and nucleoside triphosphates (Malhotra et al., 2013). However, unlike viruses, C. trachomatis possesses both DNA and RNA, possess their own ribosomes and replicate through binary fission, therefore they are known to be a bacterium (Becker et al., 1996). C. trachomatis is a powerful immunogen; meaning it is bound to components of the immune system, this stimulates both humeral and cell immune responses. The outcome associated with C. trachomatis really depends on the volume and interaction of cytokines secreted by lymphocytes (Mascellino, Boccia and Oliva, 2011).
Figure 1: coloured Transmission electron micrograph of Chlamydia trachomatis (light purple/ dark purple) inside an inclusion (white) that is inside a cell (Biomedical imaging unit, Southampton General hospital, 2013).
Chlamydia trachomatis: Pathogenesis
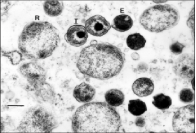
Chlamydia trachomatis has a specialised biphasic developmental cycle, C. trachomatis alternates between two morphological stages the elementary body and the reticulate body (figure 2), the bacteria throughout its life cycle alternate between the two morphological stages (Malhotra et al., 2013). The first form, the elementary body, is an extracellular form that is unable to replicate or reproduce (AbdelRahman, Belland, 2005). The elementary body is responsible for the spreading of the bacteria as they have the ability to attach and occupy susceptible cells within the host (AbdelRahman, Belland, 2005). Once the invasion of the elementary body is initialised, binding to the host cell requires several bacterial ligands (Häcker, 2018). Upon infection pre-synthesised T3SS effectors are inserted and the elementary body is endocytosed. The inclusion dissociates itself from the canonical endolysosomal pathway before differentiating into the reticulate body (Häcker, 2018); an intracellular, osmotically unstable and metabolically active form, meaning the reticulate body is unable to infect the host but is responsible for the replication of C. trachomatis undergoing multiple cycles of binary fission (Elwell, Mirrashidi and Engel, 2016). Newly secreted inclusion membrane proteins encourage the redirection of exocytic vesicles in transit from the host’s Golgi apparatus facilitating interactions between the host and pathogen (Elwell, Mirrashidi and Engel, 2016). The inclusion is then transported to the centrosome. Half way through the cycle the reticulate bodies will rapidly replicate and will secrete additional effectors that alter processes in the host cell (Elwell, Mirrashidi and Engel, 2016). During the final stages of infection, before returning to its original form, the reticulate body secretes the last of the effectors and synthesises the specific effectors associated with the elementary body, the cycle is complete when the reticulate body differentiates back to its original form the elementary body, as the elementary body is released the cycle repeats itself (Häcker, 2018).
Figure 2: transmission electron micrograph of a chlamydial infection in the liver of Xenopustropicalis. The reticulate bodies presented as R and highly condensed elementary bodies as E. 270 nm (Reed, 2000)
Gene sequencing
 The gene sequence of Chlamydia trachomatis is composed of 1,042,519 base pair chromosomes containing around 58.7% A and T and 7493 base pair plasmids (Stephens et al, 1995). Strains of C. trachomatis is classified in to serovars based on the composition of their major outer membrane protein (Seth-Smith et al., 2013), representing a major surface antigen, the serovars D-K and L1, L2 and L3 are all responsible for sexually transmitted diseases. serovars D-K are responsible for cervicitis in females and urogenital infections in males, L1, L2 and L3 are all responsible for lymphogranuloma vernerum (Thomson et al., 2007).
The gene sequence of Chlamydia trachomatis is composed of 1,042,519 base pair chromosomes containing around 58.7% A and T and 7493 base pair plasmids (Stephens et al, 1995). Strains of C. trachomatis is classified in to serovars based on the composition of their major outer membrane protein (Seth-Smith et al., 2013), representing a major surface antigen, the serovars D-K and L1, L2 and L3 are all responsible for sexually transmitted diseases. serovars D-K are responsible for cervicitis in females and urogenital infections in males, L1, L2 and L3 are all responsible for lymphogranuloma vernerum (Thomson et al., 2007).
References
- AbdelRahman, Y. and Belland, R. (2005). The chlamydial developmental cycle, FEMS Microbiology Reviews, [online] 29(5), pp.949-959. Available at: https://pdfs.semanticscholar.org/cf3c/e8fdfd0d45036a0e7b332dd75083a726b9c3.pdf [Accessed 5 Feb. 2019].
- Becker, Y. et al (1996). Medical Microbiology. 4th ed. Galveston, Texas: University of Texas Medical Branch, chapter 39.
- Biomedical imaging unit, Southampton General hospital (2013). Chlamydia trachomatis, TEM. [image] Available at: https://fineartamerica.com/featured/chlamydia-trachomatis-bacteria-tem-biomedical-imaging-unit-southampton-general-hospital.html [Accessed 13 Feb. 2019].
- Elwell, C., Mirrashidi, K. and Engel, J. (2016). Chlamydia cell biology and pathogenesis. Nature Reviews Microbiology, [online] 14(6), pp.385-400. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4886739/ [Accessed 13 Feb. 2019].
- Häcker, G. (2018). Biology of Chlamydia. 1st ed. Cham, Switzerland: Springer International Publishing, pp.3-15.
- Malhotra, M., Sood, S., Mukherjee, A., Muralidhar, S. and Bala, M. (2013). Genital Chlamydia trachomatis: An update. Indian journal of medical research, [online] vol. 138(3), pp.303-316. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3818592/ [Accessed 5 Feb. 2019].
- Mascellino, M., Boccia, P. and Oliva, A. (2011). Immunopathogenesis in Chlamydia trachomatis Infected Women. ISRN Obstetrics and Gynecology, [online] 2011, pp.1-9. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3236400/ [Accessed 10 Feb. 2019].
- Oriel, J. and Hayward, A. (1974). Sexually-transmitted diseases in animals. Sexually Transmitted Infections, [online] vol. 50(6), pp.415-416. Available at: https://www-ncbi-nlm-nih-gov.plymouth.idm.oclc.org/pmc/articles/PMC1045078/?page=4 [Accessed 5 Feb. 2019].
- Reed, K. (2000). Chlamydia infection in a breeding colony of African Clawed frogs (Xenopus tropicalis). [image] Available at: https://www.researchgate.net/publication/12560569_Chlamydia_pneumoniae_infection_in_a_breeding_colony_of_African_Clawed_frogs_Xenopus_tropicalis [Accessed 13 Feb. 2019].
- Seth-Smith, H., Harris, S., Skilton, R., Radebe, F., Golparian, D., Shipitsyna, E., Duy, P., Scott, P., Cutcliffe, L., O’Neill, C., Parmar, S., Pitt, R., Baker, S., Ison, C., Marsh, P., Jalal, H., Lewis, D., Unemo, M., Clarke, I., Parkhill, J. and Thomson, N. (2013). Whole-genome sequences of Chlamydia trachomatis directly from clinical samples without culture. Genome Research, [online] 23(5), pp.855-866. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3638141/ [Accessed 16 Feb. 2019].
- Stephens, R., Kalman, S, Lammel, C, Fan, J, Marathe, R, Aravind, L, Mitchell, W, Olinger, L, Tatusov, R, Zhao, Q, Koonin, E,. Davis, R (1998). Genome Sequence of an Obligate Intracellular Pathogen of Humans: Chlamydia trachomatis. Science, [online] 282(5389), pp.754-759. Available at: http://science.sciencemag.org/content/sci/282/5389/754.full.pdf [Accessed 14 Feb. 2019].
- Thomson, N., Holden, M., Carder, C., Lennard, N., Lockey, S., Marsh, P., Skipp, P., O’Connor, C., Goodhead, I., Norbertzcak, H., Harris, B., Ormond, D., Rance, R., Quail, M., Parkhill, J., Stephens, R. and Clarke, I. (2007). Chlamydia trachomatis: Genome sequence analysis of lymphogranuloma venereum isolates. Genome Research, [online] 18(1), pp.161-171. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2134780/ [Accessed 16 Feb. 2019].
- Yasser M. AbdelRahman, Robert J. Belland (2005) The chlamydial developmental cycle, FEMS Microbiology Reviews, [online] Vol. 29(5), pp. 949-959. Available at: https://academic.oup.com/femsre/article/29/5/949/549012#90535162 [Accessed 6 Feb. 2019]
Mate Choice
Mate choice may be defined as ‘Any pattern of behaviour shown by members of one sex that leads to there being more likely to mate with certain members of the opposite sex than with others.’ (Bateson, 1983)
Sexual selection
Sexual selection, one of Darwin’s second insights proposed 150 years ago (Jones & Ratterman, 2009) is defined as ‘The advantage at which certain individuals have over other individuals of the same and sex and species solely in respect of reproduction’ (Hosken & House, 2011). Since, other scientists studying the topic of sexual selection have added that ‘Sexual selection arises from differences in reproductive success caused by competition for access to mates’ (Jones & Ratterman, 2009)
Sexual selection drives an asymmetric introgression, which is the movement of a gene from one species into the gene pool of another (Garcia-Roa & Carazo, 2017).
It was realised by Darwin that sexual selection could be arbitrated by male–male combat or by a female’s choice of attractive males with phenotypic benefits. (Jones & Ratterman, 2009)
Intra-sexual reproduction
Intra-sexual reproduction is where members of the same sex compete for mates. There is a strong selection for large body size and weapons, which are linked to the hierarchies of male dominance. In species where males establish dominance relationships with one another, females may benefit to mate preferentially with a male of high status. This creates advantages to their direct phenotypic benefits, sensory biases and indicator traits (Bateson, 1983).
Dominant males have the best direct phenotypic benefits, sensory biases and indicator traits. They usually have larger and higher quality habitats and territories, meaning that they have better access to females, better parental care and access to food. The success of fertilisation through being able to mate without interruption, the father’s parental protection and also genetic advantages for the offspring status.
The success of fertilisation is strongly linked to the dominance of the males and how well they compete for a mate. (Jacob et al, 2009)
Inter-sexual reproduction
Inter-sexual reproduction is an evolutionary process which selection of a mate is dependent on the attractiveness of phenotypic traits. In most but not all cases females choose against competing males.

The inter-sexual indicator model
The dark grey ovals represent the different types of inter-sexual selection:
A) female preference for male signals
B) the correlation between male quality and male signals
C) female preference for underlying male quality.
The three light grey bands that join the ovals signify the three relationships that are involved in inter-sexual selection. The thicknesses of the bands represent the predicted relative strengths of these phenotypic relationships. The straight horizontal bands are direct relationships where one trait is directly related to the other, conversely, the curved band is an indirect relationship where the two traits are linked by an intermediate ‘signal’ trait.
Signal traits
Phylogenetic signal is the tendency for closely related species to show similar trait values as a consequence of their phylogenetic proximity.
Cite This Work
To export a reference to this article please select a referencing style below:

